SecureEpiLinker (SEL) S. Stammler, T. Kussel, P. Schoppmann, F. Stampe, G. Tremper, S. Katzenbeisser, K. Hamacher, M. Lablans. Mainzelliste SecureEpiLinker (MainSEL): Privacy-Preserving Record Linkage using Secure Multi-Party Computation, Bioinformatics, 38(6): 1657-1668, 2020 Paper Code on GitHub | |
BioTite P. Kunzmann, K. Hamacher. Biotite: A unifying open source computational biology framework in Python, BMC Bioinformatics, 19:346, 2018. Code on GitHub | streaMD R package to analyze MD trajectories in a graph-semantic fashion R package to analyze MD trajectories in a graph-semantic fashion J. Comp. Chem., 39:1666-1674, 2018. Abstract & PDF-Version Cover Page |
TransferEntropyPT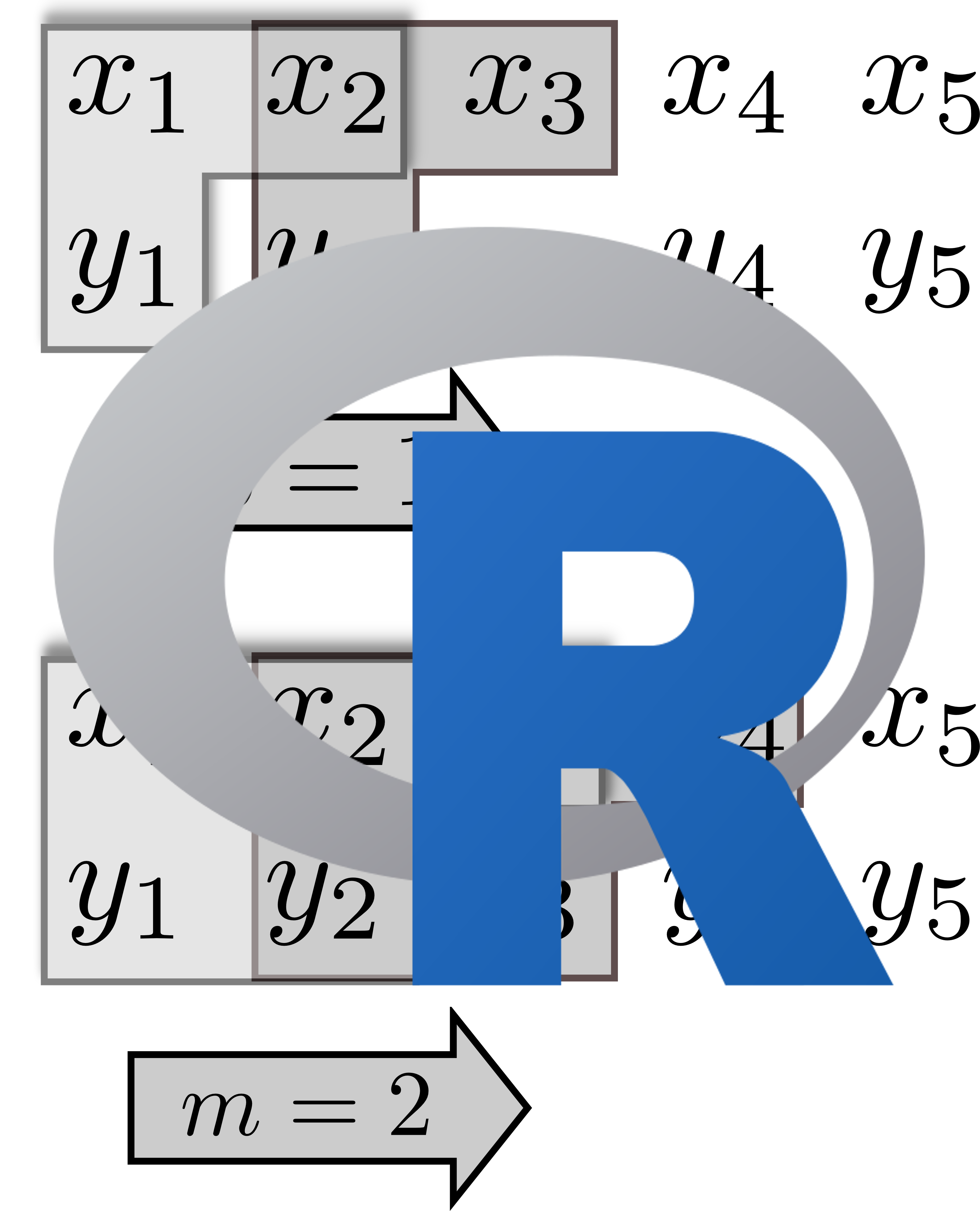 R package to compute transfer entropy (under statistical permutation test) R package to compute transfer entropy (under statistical permutation test) 15th conference on Computational Methods for Systems Biology Lecture Notes in Bioinformatics (LNBI 10545), 285-290, 2017. Abstract & PDF-Version | DOCKTITE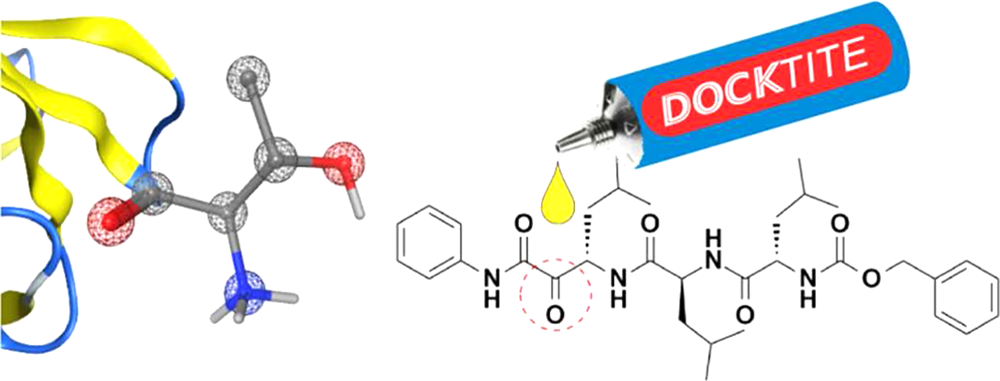 Efficient, reliable covalent docking in Molecular Operating Environment (MOE) Efficient, reliable covalent docking in Molecular Operating Environment (MOE)SVL-Source-Code at MOE repository Subscribe to Newsletter J. Chem. Inf. Model. 55(2):398-406, 2015. |
Resampling in Lisp a domain-specific language (DSL) for statistical tests of arbitrary S-expressions (and more) Source code (tar.gz) | BioPhysConnectoR Molecular (Co)evolution and Biophysical/Molecular Dynamics in R; Molecular (Co)evolution and Biophysical/Molecular Dynamics in R;uses Information Theory, Elastic Network Models, Multi-Core Computations etc. Code at CRAN Our BioPhysConnectoR-Website BMC Bioinformatics 2010, 11:199 |
libTransferEntropy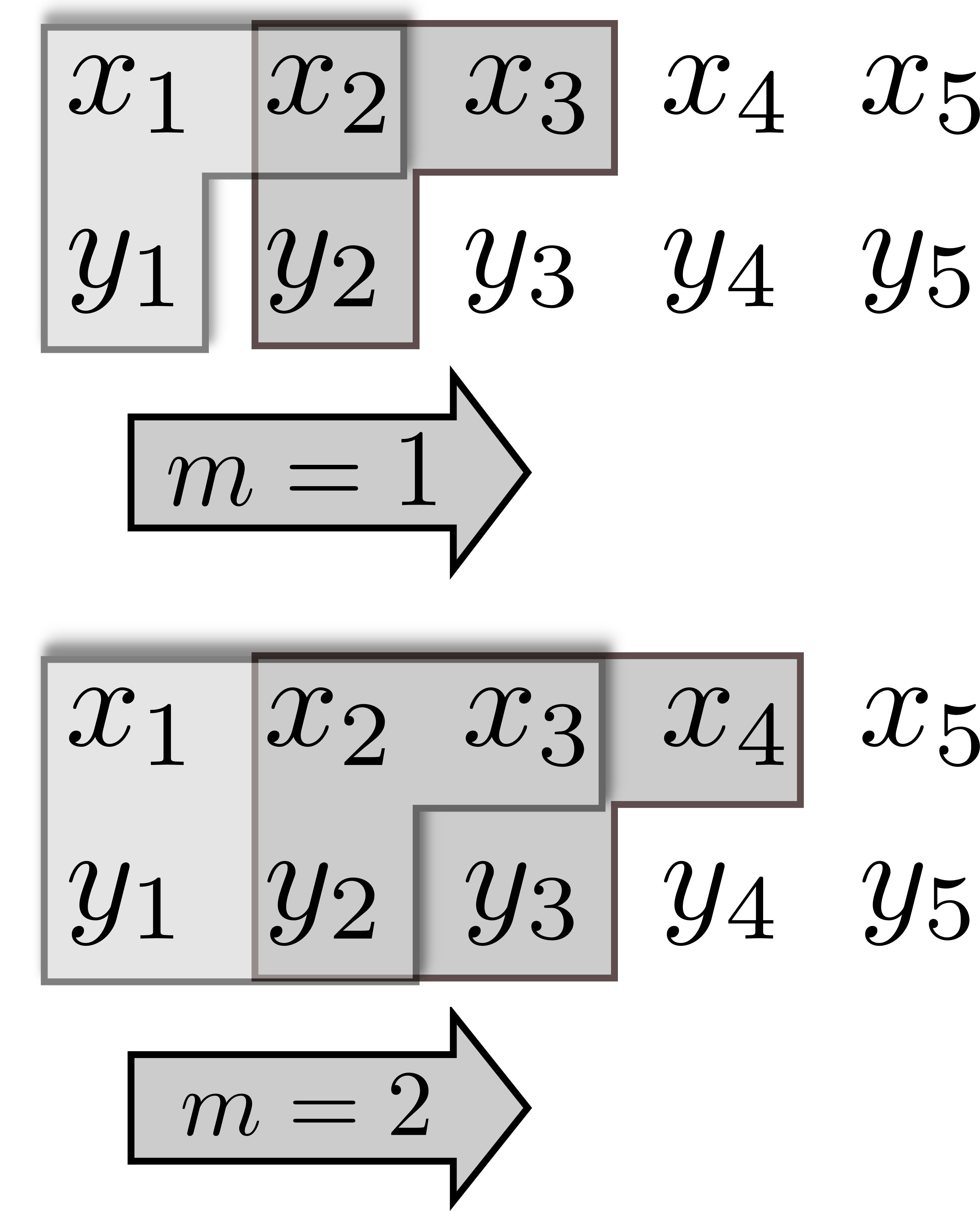 Compute transfer entropy (including statistical permutation test) for time series Frontiers in Physics 3(2015)10 Compute transfer entropy (including statistical permutation test) for time series Frontiers in Physics 3(2015)10 | ViPhyVisual Analytics for PhylogeniesViPhy-Website IEEE Conference on Visual Analytics Science and Technology (VAST2011), 31-40, 2011 |
CoMIC - Coevolution via MI on CUDA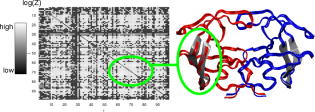 We've implemented a massively-parallel version of our normalization procedure for mutual information values in molecular (co)evolution. To this end we compute replica-wise -scores and percentiles of a parameter-free model of neutral evolution. We've implemented a massively-parallel version of our normalization procedure for mutual information values in molecular (co)evolution. To this end we compute replica-wise -scores and percentiles of a parameter-free model of neutral evolution.CoMIC-Website | MatrixVisVisual Analytics for (Co)evolutionary MatricesMatrixVis-Website BMC Bioinformatics, 11:330, 2010 |